questioner121 wrote:I think I'm beginning to understand
The content of your posts says otherwise.
questioner121 wrote:where the evolutionists
Oh no, it's that tiresome creationist canard ...
The "evolutionist" canard (with "Darwinist" side salad).Now, if there is
one guaranteed way for a creationist to establish that he or she is here for no other reason than to propagandise for a doctrine, it's the deployment of that most
viscerally hated of words in the lexicon, namely,
evolutionist. I have posted about this so often here, that I was surprised to find that I'd missed it out of the original list, but I had more pressing concerns to attend to when compiling the list originally. However, having been reminded of it, now is the time to nail this one to the ground with a stake through its heart once and for all.
There is
no such thing as an "evolutionist". Why do I say this? Simple. Because the word has become thoroughly
debased through creationist abuse thereof, and in my view, deserves to be struck from the language forever. For those who
need the requisite education, there exist
evolutionary biologists, namely the scientific professionals who devote decades of their lives to understanding the biosphere and conducting research into appropriate biological phenomena, and those outside that specialist professional remit who accept the reality-based, evidence-based case that they present in their peer reviewed scientific papers for their postulates. The word "evolutionist" is a
discoursive elision, erected by creationists for a very specific and utterly mendacious purpose, namely to suggest that valid evolutionary science is a "doctrine", and that those who accept its postulates do so merely as
a priori "assumptions" (see [3] above). This is
manifestly false, as anyone who has actually
read the peer reviewed scientific literature is eminently well placed to understand. The idea that there exists some sort of "symmetry" between valid, evidence-based, reality-based science (evolutionary biology) and assertion-laden, mythology-based doctrine (creationism) is
FALSE. Evolutionary biology, like every other branch of science,
tests assertions and presuppositions to destruction, which is why creationism was tossed into the bin 150 years ago (see [2] above). When creationists can provide methodologically rigorous empirical tests of their assertions, the critical thinkers will sit up and take notice.
Furthermore, with respect to this canard, does the acceptance of the scientifically educated individuals on this board, of the current scientific paradigm for gravity make them "gravitationists"? Does their acceptance of the evidence supporting the germ theory of disease make them "microbists"? Does their acceptance of the validity of Maxwell's Equations make them "electromagnetists"? Does their acceptance of of the validity of the work of Planck, Bohr, Schrödinger, Dirac and a dozen others in the relevant field make them "quantumists"? Does their acceptance of the validity of the astrophysical model for star formation and the processes that take place inside stars make them "stellarists"? If you are unable to see the absurdity inherent in this, then you are in no position to tell people here that professional scientists have got it wrong, whilst ignorant Bronze Age nomads writing mythology 3,000 years ago got it right.
While we're at it, let's deal with the duplicitous side salad known as "Darwinist". The critical thinkers here
know why this particular discoursive elision is erected, and the reason is related to the above. Basically, "Darwinist" is erected for the specific purpose of suggesting that the only reason people accept evolution is because they bow uncritically to Darwin as an authority figure. This is, not to put too fine a point on it, droolingly encephalitic drivel of a particularly suppurating order. Let's provide a much needed education once and for all here.
Darwin is regarded as
historically important because he founded the scientific discipline of evolutionary biology, and in the process, converted biology from a cataloguing exercise into a proper empirical science. The reason Darwin is considered important is NOT because he is regarded uncritically as an "authority figure" - the critical thinkers leave this sort of starry-eyed gazing to followers of the likes of William Lane Craig. Darwin is regarded as important because
he was the first person to pay serious attention to reality with respect to the biosphere, with respect to the business of determining mechanisms for its development, and the first to engage in diligent intellectual labour for the purpose of establishing that reality supported his postulates with respect to the biosphere. In other words, instead of sitting around accepting uncritically mythological blind assertion, he got off his arse, rolled up his sleeves, did the hard work, put in the long hours performing the research and gathering the real world data, and then spending long hours determining what would
falsify his ideas and determining in a rigorous manner that no such falsification existed. For those who are unaware of this, the requisite labour swallowed up
twenty years of his life, which is par for the course for a scientist introducing a new paradigm to the world.
THAT is why he is regarded as important, because
he expended colossal amounts of labour ensuring that REALITY supported his ideas. That's the ONLY reason ANY scientist acquires a reputation for being a towering contributor to the field, because said scientist toils unceasingly for many years, in some cases
whole decades, ensuring that his ideas are supported by reality in a methodologically rigorous fashion.
Additionally, just in case this idea hasn't crossed the mind of any creationist posting here, evolutionary biology has moved on in the 150 years since Darwin, and whilst his historical role is rightly recognised, the critical thinkers have also recognised that more recent developments have taken place that would leave Darwin's eyes out on stalks if he were around to see them. The contributors to the field
after Darwin are
numerous, and include individuals who contributed to the development of other branches of science making advances in evolutionary theory possible. Individuals such as Ronald Fisher, who developed the mathematical tools required to make sense of vast swathes of biological data (heard of analysis of variance? Fisher invented it), or Theodosius Dobzhansky, who combined theoretical imagination with empirical rigour, and who, amongst other developments, provided science with a documented instance of speciation in the laboratory. Other seminal contributors included Müller (who trashed Behe's nonsense six decades before Behe was born), E. O. Wilson, Ernst Mayr, Motoo Kimura, Stephen Jay Gould, Niles Eldredge, J. B. S. Haldane, Richard Lewontin, Sewall Wright, Jerry Coyne, Carl Woese, Kenneth Miller, and they're just the ones I can list off the top of my head. Pick up any half-decent collection of scientific papers from the past 100 years, and dozens more names can be added to that list.
So, anyone who wants to be regarded as an extremely low-grade chew toy here only has to erect the "evolutionist" or "Darwinist" canard, and they will guarantee this end result.
Read the above and learn from it.
questioner121 wrote:on this forum are coming from. They assume
Wrong again. We don't "assume" anything. We leave assumptions, and for that matter, blind assertions and presuppositions, to supernaturalists.
questioner121 wrote:that variations in animals is natural
It is. Are you identical to either of your parfents? No? There's part of your evidence. Moreover, if you pick up an actual biology textbook, and look up meiosis, you'll find
reasons for the emergence of variation in offspring (at least, in sexually reproducing organisms) arising from the mechanism by which genes are shuffled when germ cells are made.
questioner121 wrote:so it's perfectly possible to have a mutations which give birds a navigation system, for zebras to have stripes, for peacocks to have extravagant feathers, etc.
And lo and behold, we have
evidence for countless thousands of mutations. I'm aware of several mutations in just one species of tropical fish that have appeared during the 100 years or so of its domestication in the aquarium. What's more, many of the genes in question are known to obey well-defined inheritance mechanisms. The Double Tail mutation in
Betta splendens is autosomal recessive, and obeys the Mendelian laws of inheritance for single-factor mutations.
questioner121 wrote:The only thing I'm unclear is on are these mutations big bang or did they develop gradually over millions of years?
Actually, any mutation (and it has to be a mutation in a germ cell, in order for it to be passed on to offspring) can occur at any moment. What takes time is the
fixation of a mutation in a population, once that mutation has been disseminated to offspring. Which depends upon the strength of the selection pressure leading to fixation thereof. Mutations that are subject to strong selection pressures will achieve fixation in a population in a relatively small number of generations (usually, one is looking at somewhere between 50 and 100 generations for this to take place). If the organism in question has a fast turnaround, and produces a new generation every month, then fixation can occur in less than 5 years if the selection pressure is long enough, If, on the other hand, the organism in question takes 20 years to produce a new generation, then fixation won't occur until after something like 2,000 years has elasped, and that's if we're dealing with a strong selection pressure. A weak selection pressure will increase the time for each case.
questioner121 wrote:Did bird migration start of with a basic navigation system, did the zebras start of having grey stripes which deepened to black, did the peacock start off having 1 feather which then became 100+?
Are you incapable of finding this out for yourself? Google Scholar will point you to thousands of scientific papers if you bother to use it.
questioner121 wrote:I think gene mutations are 'natural' but for a gene mutation to actually develop a well functioning trait is something that is unusual to say the least.
No it isn't. Do I have to bring the Double Tail mutation in
Betta splendens here to demonstrate this?
questioner121 wrote:If you look at the genetic level, a mutation to create, let's say for example, an extra digit on a persons hand doesn't just mean creating an extra finger but also wiring it up to the brain, plugging it into the blood circulation system, maintaining it, etc.
So what? This is familiar stuff to those who bothered to learn about regulatory networks and evo-devo. You'll also find that there are quite a few humans on the planet with extra digits (a condition known as polydactyly). Here's an x-ray of a human foot with 6 toes, for example:

Here's an x-ray of a human hand with 6 fingers:
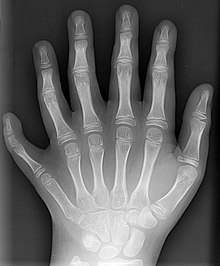
Plus, polydactyly is observed fairly frequently in various cat breeds.
Here's a news item featuring a polydactyl cat. Cats normally have five toes on the front paws, and four on the rear. This cat has 7 on the front, and 6 on the rear. All of them fully operational. Indeed, in the past, polydactyl cats were regarded as not only being omens of good luck amongst sailors in the days of sailing ships, but were valued for their enhanced climbing ability and rodent hunting skills.
None of this is a problem for those of us who bothered to study actual biology.
questioner121 wrote:I can understand it if evolutionists
Yawn. Read my above destruction of the tiresome "evolutionist" canard.
questioner121 wrote:think it's perfectly naturally for animals in a population to have an extra digit here or there, different coloured skins, different shaped beaks, etc. But I don't think evolutionists understand that it's not a simple matter of a genetic mutation at the molecular level.
The evidence says otherwise. The Double Tail mutation in
Betta splendens is centred upon a single gene, one that exhibits classic Mendelian inheritance.
questioner121 wrote:It needs to encompass a significant proportion of the animals DNA in order to make the genetic mutation viable.
Wrong. Do I have to bring the Double Tail mutation in
Betta splendens here, in order to establish that you are wrong?
questioner121 wrote:ramseyoptom wrote:How can you be unclear on how the mutations came about, have you not read any of what has been posted or the links to other information?
Sorry I didn't see any link which describes how a mutation develops at the genetic level. The links just pointed to what mutations are observed in nature and then whole load of speculation as to how they could have been naturally selected for.
Oh please, go and pick up an actual science textbook. Scientists have catalogued
numerous chemical reactions that result in mutations. The classic example being methylation of a cytosine molecule in a DNA strand, to form 5-methylcytosine, followed by deamination of that molecule, which results in the molecule changing to thymine, a different chemical base. A C->T change in a DNA strand can have profound effects. For example, changing a C to a T in a CGA codon (which normally codes for the amino acid arginine) results in the codon becoming a stop codon, and signalling termination of protein synthesis. Change the C in a CTT codon (normally coding for leucine) to a T (resulting in a TTT codon, coding for phenylalanine) and the resulting protein could have a significantly altered function, depending upon whether or not that amino acid is in a critical region.
Then of course, there are other classes of mutations apart from point mutations, such as insertion and deletions (indels), frameshift mutations (responsible for nylonase in Japanese
Flavobacterium) and whole gene duplications (which resulted in the emergence of antifreeze glycoproteins in Antarctic Notothenioid fishes, when the duplicated trypsinogen gene acquired mutations that resulted in the protein coded for possessing a new function).
questioner121 wrote:Basically the entire explanation on the diversity of life from 'simple' cell organisms to multi cell organisms relies on mutations to present within any given population. And that's it. Nothing else. The direction of the diversity of organism population is then based on NS or some other mechanism.
You do realise we have
evidence for the emergence of multicellularity from single celled organisms? It's been observed in the laboratory, and the effect can be produced releatably and reliably in laboratory experiments.
questioner121 wrote:So when the evolutionists
Yawn.
questioner121 wrote:reads thousands of papers on observations of genetic mutations and how they are selected for it immediately tells them that this is what happened 'billions' of years ago.
This might have something to do with the fact that we have
zero evidence for the chemistry of life being different in the past. Indeed, if that chemistry
was different in the past, then
we would have inherited that chemistry. You do know what "inheritance" means, don't you?
questioner121 wrote:Now that is just plain ridiculous.
No it isn't. It makes perfect sense to postulate that we inherited the same basic chemistry as the earliest life forms. Unless you can provide a detailed exposition of how a
different chemistry could have been inherited, of course ...
questioner121 wrote:How can someone make such a huge assumption?
It isn't an "assumption". How do we know this? Oh, that's right, various experiments have been conducted in the laboratory aimed at elucidating how that chemisttry arose. How many of the 116 or sso papers on this do you want me to bring here?
questioner121 wrote:And then to go on and trumpet about how humans came from apes as a fact is even crazier.
Oh dear, you really never attended any science classes, did you?
I have news for you.
Linnaeus, way back in 1747, thought humans and chimpanzees were suffficiently closely related, on the basis of comparative anatomy alone, for the two species to belong to the same taxonomic Genus. The reason he didn't do this was
interference from religion, in an era when enforcers of conformtiy to doctrine had the power to destroy your life. Indeed, I've covered this in the past here on numerous occasions: Linnaeus wrote a letter to his fellow taxonomist, Johann Georg Gmelin, in 1747, lamenting this interference. The letter can be read in full online, courtesy of the University of Uppsala's publicly available version of the database containing the letters of Linnaeus
here. The letter in question is
this one, dated 25th February, 1747. I'll provide both the original Latin text of the relevant paragraph, and the English translation:
Linnaeus, 1747 wrote:Non placet, quod Hominem inter ant[h]ropomorpha collocaverim, sed homo noscit se ipsum. Removeamus vocabula. Mihi perinde erit, quo nomine utamur. Sed quaero a Te et Toto orbe differentiam genericam inter hominem et Simiam, quae ex principiis Historiae naturalis. Ego certissime nullam novi. Utinam aliquis mihi unicam diceret! Si vocassem hominem simiam vel vice versa omnes in me conjecissem theologos. Debuissem forte ex lege artis.
The translation reads as follows:
It does not please (you) that I've placed Man among the Anthropomorpha,[22] but man learns to know himself. Let's not quibble over words. It will be the same to me whatever name we apply. But I seek from you and from the whole world a generic difference between man and simian that [follows] from the principles of Natural History. I absolutely know of none. If only someone might tell me a single one! If I would have called man a simian or vice versa, I would have brought together all the theologians against me. Perhaps I ought to have by virtue of the law of the discipline.
So Linnaeus, who was a
de facto creationist (purely because there existed no other paradigm in his day), agreed that humans and chimpanzees were sufficiently closely related, to warrant their placement in the same taoxnomic Genus, yet was inhibited from doing so by
religious interference in science. Of course this is not the only instance where the attitude of the religious has consisted of "conform or else", not is it the only instance of the religious insisting that doctrine is automatically right, regardless of whether or not reality agrees with this. But I digress. The simple fact is, comparative anatomists,
creationists included, throughout the 18th and early 19th centuries, posited that
relationships existed between different organismal taxa, on the basis of shared anatomical characteristics.
The master stroke Darwin provided, was to provide a
mechanism for the appearance of those shared anatomical characteristics. Namely,
inheritance from shared ancestors. Indeed, inheritance is of seminal importance throughout the early Abrahamic mythologies, which regale us with an abundance of tedious genealogical lists. Darwin's contribution consisted of extending genealogy across the entire biosphere. It's not as if we lack
evidence for the existence of inheritance followed by divergence: every instance of selective breeding performed by humans, in pursuit first of ensuring a food supply via agriculture, provides evidence for this mechanism in action. There is
no evidence whatsoever, that the relevant genetic mechanisms underpinning this operated in different ways in the past. Consequently, since we have
evidence for inheritance followed by acquisition of new features (including documented instances of speciation in the scientific literature), it is entirely apposite to postulate that the same processes applied to organisms in the past, including fossil organisms. And, when those fossil organisms are subject to the
same analysis of comparative anatomy applied to living organisms, we find patterns of the same class appearing again and again - common anatomical features being dispensed to later generations, then subject to stepwise modification.
The point being made here, of course, is that
even if no fossils had ever been found, the evidence from living organisms is sufficient to make the case for evolution, because the analysis of those fossils derives from work performed on present day living organisms. Fossils are simply the icing on the cake. Even more importantly, when the underlying biochemical basis for genetics was alighted upon, the potential existed for that discovery to
falsify evolutionary postulates on a grand scale. But oh, that's right, it
didn't falsify evolutionary postulates, it
reinforced them. Hence the Modern Synthesis.
Oh, and the evidence from the genomes of the two species merely reinforces this.
Let's take a look at a relevant scientific paper, shall we? Namely this one:
Initial Sequencing Of The Chimpanzee Genome And Comparison With The Human Genome, The Chimpanzee Genome Sequencing Consortium (see paper for full list of 68 authors),
Nature,
437: 69-87 (1 September 2005) [Full paper downloadable from
here]
Chimpanzee Genome Sequencing Consortium wrote:Here we present a draft genome sequence of the common chimpanzee (Pan troglodytes). Through comparison with the human genome, we have generated a largely complete catalogue of the genetic differences that have accumulated since the human and chimpanzee species diverged from our common ancestor, constituting approximately thirty-five million single-nucleotide changes, five million insertion/deletion events, and various chromosomal rearrangements. We use this catalogue to explore the magnitude and regional variation of mutational forces shaping these two genomes, and the strength of positive and negative selection acting on their genes. In particular, we find that the patterns of evolution in human and chimpanzee protein-coding genes are highly correlated and dominated by the fixation of neutral and slightly deleterious alleles. We also use the chimpanzee genome as an outgroup to investigate human population genetics and identify signatures of selective sweeps in recent human evolution.
Let's take a look at the detailed findings, shall we?
Chimpanzee Genome sequencing Consortium wrote:Here we report a draft sequence of the genome of the common chimpanzee, and undertake comparative analyses with the human genome. This comparison differs fundamentally from recent comparative genomic studies of mouse, rat, chicken and fish
14–17. Because these species have diverged substantially from the human lineage, the focus in such studies is on accurate alignment of the genomes and recognition of regions of unusually high evolutionary conservation to pinpoint functional elements. Because the chimpanzee lies at such a short evolutionary distance with respect to human, nearly all of the bases are identical by descent and sequences can be readily aligned except in recently derived, large repetitive regions. The focus thus turns to differences rather than similarities. An observed difference at a site nearly always represents a single event, not multiple independent changes over time. Most of the differences reflect random genetic drift, and thus they hold extensive information about mutational processes and negative selection that can be readily mined with current analytical techniques. Hidden among the differences is a minority of functionally important changes that underlie the phenotypic differences between the two species. Our ability to distinguish such sites is currently quite limited, but the catalogue of human–chimpanzee differences opens this issue to systematic investigation for the first time.We would also hope that, in elaborating the few differences that separate the two species, we will increase pressure to save chimpanzees and other great apes in the wild.
Our results confirm many earlier observations, but notably challenge some previous claims based on more limited data. The genome-wide data also allow some questions to be addressed for the first time. (Here and throughout, we refer to chimpanzee–human comparison as representing hominids and mouse–rat comparison as representing murids—of course, each pair covers only a subset of the
clade.) The main findings include:
. Single-nucleotide substitutions occur at a mean rate of 1.23% between copies of the human and chimpanzee genome,
with 1.06% or less corresponding to fixed divergence between the species.
. Regional variation in nucleotide substitution rates is conserved between the hominid and murid genomes, but rates in subtelomeric regions are disproportionately elevated in the hominids.
. Substitutions at CpG dinucleotides, which constitute one-quarter of all observed substitutions, occur at more similar rates in male and female germ lines than non-CpG substitutions.
. Insertion and deletion (indel) events are fewer in number than single-nucleotide substitutions, but result in ~1.5% of the euchromatic sequence in each species being lineage-specific.
. There are notable differences in the rate of transposable element insertions: short interspersed elements (SINEs) have been threefold more active in humans, whereas chimpanzees have acquired two new families of retroviral elements.
.
Orthologous proteins in human and chimpanzee are extremely similar, with ~29% being identical and the typical orthologue differing by only two amino acids, one per lineage.
. The normalized rates of amino-acid-altering substitutions in the hominid lineages are elevated relative to the murid lineages, but close to that seen for common human polymorphisms, implying that positive selection during hominid evolution accounts for a smaller fraction of protein divergence than suggested in some previous reports.
. The substitution rate at silent sites in exons is lower than the rate at nearby intronic sites, consistent with weak purifying selection on silent sites in mammals.
. Analysis of the pattern of human diversity relative to hominid divergence identifies several loci as potential candidates for strong selective sweeps in recent human history.
So, according to the
actual research scientists who analysed the chimpanzee genome, it has been found that
around twenty nine percent of the orthologous proteins of the two species are IDENTICAL, with the remainder differing by just one or two amino acids. This finding
only makes sense, along with several other findings, if one postulates that the genes in question were inherited from a common ancestor. I'll deal with retroviral insertions in a moment. But first, I want to deal with another matter, namely the work on human chromosome 2, which has been found to be the result of a chromosome fusion event in the past, after the human and chimpanzee lineages split from our common ancestor. Here's evolutionary biologist Ken Miller explaining the findings in a video clip - I'll post the full transcript after the video:
[youtube]http://www.youtube.com/watch?v=Gs1zeWWIm5M[/youtube]
Ken Miller wrote:The second thing that you saw at the trial, was that when data was introduced at the trial, which I and another witness introduced from whole genome sequencing,
the intelligent design advocates just literally had nothing to say. We weren't asked questions in cross-examination, the other side never brought it up, they never argued against it, they just left it. Here's an example.
Many of you may know that a few months ago the genetic code of the chimpanzee was published. Therefore we can compare our genome to these primate relatives. What do we find? I want to show you one striking finding that dates to about a year ago. You all know that evolution argues that we share a common ancestor with the great apes - the chimpanzee, the gorilla and the orang-utan. Well, if that's true there should be genetic similarities, and in fact there are.
But there's something that's really interesting, and has the potential, if it were true, to contradict evolutionary common ancestry, and that is we have two fewer chromosomes than the other great apes - we have 46, they all have 48. That's very interesting. Now what does that actually mean? Well first of all, the 46 chromosomes that we have - you've got 23 from Mom and 23 from Dad, so it's actually 23
pairs - these guys have 24 from each parent so they have 24 pairs. So everybody in this room is
missing a pair of chromosomes, so where did it go? Could if have gotten lost in our lineage? Ah-ah - if it got lost, if a whole primate chromosome was lost, that would be lethal. So there's only two possibilities, and that is if these guys
really share a common ancestor, that ancestor either had 48 chromosomes or 46. Now if it had 48 - 24 pairs, which is probably true, because 3 our of 4 have 48 chromosomes -
what must have happened is that one pair of chromosomes must have gotten fused. So we should be able to look at our genome, and discover that one of our chromosomes resulted from the
fusion of two primate chromosomes.
So we should be able to look around our genome, and you know what, if we don't find it, evolution is wrong - we don't share a common ancestor. So if - how would we find it?Well, biologists in the room will know that the chromosomes have nifty little markers - they have markers called
centromeres which are DNA sequences which are used to separate them during mitosis, and they have cool little DNA sequences on the ends called
telomeres. What would happen if a pair of chromosomes got fused? Well what would happen is the fusion would put telomeres
where they don't belong in the centre of the chromosome, and the resulting fused chromosome should actually have
two centromeres. One of them might become inactivated, but nonetheless it should still be there.
So we can scan our genome, and you know that if we don't find that chromosome, evolution's in trouble.Well, guess what?
It's chromosome number 2.Our chromosome number 2 was formed by the fusion of two primate chromosomes - this is the paper from
Nature a little more than a year ago - and I put up a little of the paper, I'm sorry if it's technical but look at what it says!
"Chromosome 2 is unique to our lineage. It emerged as a result of head-to-head fusion of two chromosomes that remain separate in other primates." Those of you who have not kept up with how much we know about the genome should pay attention to this because you'll be amazed at how precisely we can look at things ...
the precise fusion site has been located at base number 114,455,823 to 114, 455, 838 ... in other words, within
fifteen bases ... and you'll notice -
multiple sub-telomere duplications - the telomeres that don't belong, and lo and behold, the centromere that is inactivated
corresponds to chimp chromosome 13. It's there, it's testable, it confirms the prediction of evolution. How would intelligent design explain this? Only one way - by shrugging and saying "that's the way the designer made it" - no reason, no rhyme, presumably there's a designer who
designed human chromosome number 2 to make it look
as if it was formed by the fusion from a primate ancestor ... I'm a Roman Catholic, I'm a theist in the broadest sense, I would say that I believe in a 'designer', but you know what, I don't believe in a
deceptive one, I don't believe in one who would do this to try to fool us, and therefore I think this is authentic - it tells us something about our ancestry.
And now, I shall return to retroviral insertions. These are snippets of viral genes, that appear in our genome, arising from the action of various retroviruses, courtesy of the interesting and unusual manner in which these viruses reproduce in host cells. Since the genomes of retroviruses contain many common features, including the ubiquitous presence of genes known as
gag, pol and
env, appearance of fragments of these genes indicate the past action of a retrovirus.
During their reproduction, retroviruses insert their genomes into the DNA of the host cells, and in effect, hijack the host cells' transcription and translation machinery to do the work of building new viruses. The interesting point to note here is that retroviruses are not especially discriminating about
where in the genome of the host cell they insert their genetic material. They insert their genetic material into whichever location happens to be conveniently available at the time, and express relatively little preference for any particular segment of the target genome. As a result, retroviral insertions can appear
anywhere in a host genome.
Now, the fun part is this. When retroviruses attack germ cells responsible for reproduction, and leave gene fragments behind in those germ cells, those germ cells can then go on to produce sperm and eggs, that pass on those retroviral insertions to future offspring. Over the course of many generations, a species can acquire numerous different retroviral insertions, at different points in the genome, and these insertions are mostly random. Therefore, a retrovirus infecting two different species independently, will almost certainly leave behind retroviral insertions at
different places in the genomes of those two species. The probability of those insertions appearing in the
same place in the two species, under such conditions, is
extremely small. However, if a species acquires retroviral insertions, then diverges and produces two lineages that later become fully-fledged and separate species in their own right, both of those species will inherit the same pattern of retroviral insertions. Therefore, if two species share a common ancestor, one major test that can be performed, consists of searching for retroviral insertions, and seeing if those retroviral insertions line up in the two different species.
When this is performed with the human and chimpanzee genomes, what do we find? We find no less than
sixteen retroviral insertions in
identical places in the two genomes. This is a finding that ONLY makes sense, if those retroviral insertions were inherited from a common ancestor. The probability of
sixteen retroviral insertions appearing independently in two different species via separate infective events is
absurdly tiny - taking a figure of 3.5 × 10
9 base pairs for the size of the human and chimpanzee genomes, the probability of simultaneous identical retroviral insertions via separate infective events is [1/(3.5 ×10
9)]
16, which is 1.97 ×10
-153, a pobability so low that one would have to wait a
minimum of 10
154 years to see such an event happen. On the other hand, if those retroviral insertions were acquired via inheritance from a common ancestor, the probability is precisely 1.
Similar comments apply to pseudogenes (genes that once functioned, but no longer do so), though the mechanism by which these are derived is different from that applicable to retroviral insertions. However, the same pseudogene appearing in the same place in two different lineages, is again strong evidence that the two lineages in question shared a past common ancestor. The classic example in humans and chimpanzees is the GULO pseudogene, which once functioned as the gene coding for the enzyme completing the production of vitamin C. This gene no longer works in either humans or chimpanzees, and lo and behold, the manner in which the gene is broken is the
same in humans and chimpanzees. However, a broken GULO pseudogene also appears in guinea pigs, but the manner in which the gene is broken in the guinea pig lineage is
different to that seen in humans and chimpanzees, which means that the guinea pig pseudogene was broken independently long
after the distant common ancestor of the primate and rodent lineages gave rise to those lineages.
A relevant paper is this one:
Retroviral And Pseudogene Insertion Sites Reveal The Lineage Of Human Salivary And Pancreatic Amylase Genes From A Single Gene During Primate Evolution by Linda C. Samuelson, Karin Weibauer, Claudette M. Snow and Miriam H. Meisler,
Molecular and Cellular Biology,
10(6): 2513-2520 (June 1990) [Full paper downloadable from
here]
Here's the introduction from the authors:
Samuelson et al, 1990 wrote:We have analyzed the junction regions of inserted elements within the human amylase gene complex. This complex contains five genes which are expressed at high levels either in the pancreas or in the parotid gland. The proximal 5'-flanking regions of these genes contain two inserted elements.
A γ-actin pseudogene is located at a position 200 base pairs upstream of the first coding exon. All of the amylase genes contain this insert. The subsequent insertion of an endogenous retrovirus interrupted the γ-actin pseudogene within its 3'-untranslated region. Nucleotide sequence analysis of the inserted elements associated with each of the five human amylase genes has revealed a series of molecular events during the recent history of this gene family. The data indicate that the entire gene family was generated during primate evolution from one ancestral gene copy and that the retroviral insertion activated a cryptic promoter.
The above sequence of events is
extremely improbable if one assumes that humans and other great apes did not share a common ancestor. On the other hand, the above sequence of events has probability 1 if the great apes
did share a common ancestor.
Indeed, the appearance of specific patterns of retroviral insertions shared between different lineages is considered, as a result of the known mechanisms for insertions of retroviral genome fragments into a host lineage, is considered
hugely informative with respect to the pattern of common ancestry of those lineages. Here's another paper, which documents the inheritance of a pattern specific to humans and gorillas
after the human lineage split from the common ancestor:
Lineage-Specific Expansions Of Retroviral Insertions Within The Genomes Of African Great Apes But Not Humans And Orangutans by Chris T. Yohn, Zhaoshi Jiang, Sean D. McGrath, Karen E. Hayden, Philipp Khaitovich, Matthew E. Johnson, Marla Y. Eichle2, John D. McPherson, Shaying Zhao, Svante Pääbo and Evan E. Eichler,
PLoS Biology,
3(4): e110 (2005) [DOI: 10.1371/journal.pbio.0030110, full paper downloadable from
here]
Yohn et al, 2005 wrote:Retroviral infections of the germline have the potential to episodically alter gene function and genome structure during the course of evolution. Horizontal transmissions between species have been proposed, but little evidence exists for such events in the human/great ape lineage of evolution. Based on analysis of finished BAC chimpanzee genome sequence, we characterize a retroviral element (
Pan troglodytes endogenous retrovirus 1 [PTERV1]) that has become integrated in the germline of African great ape and Old World monkey species but is absent from humans and Asian ape genomes.
We unambiguously map 287 retroviral integration sites and determine that approximately 95.8% of the insertions occur at non-orthologous regions between closely related species. Phylogenetic analysis of the endogenous retrovirus reveals that
the gorilla and chimpanzee elements share a monophyletic origin with a subset of the Old World monkey retroviral elements, but that the average sequence divergence exceeds neutral expectation for a strictly nuclear inherited DNA molecule. Within the chimpanzee, there is a significant integration bias against genes, with only 14 of these insertions mapping within intronic regions. Six out of ten of these genes, for which there are expression data, show significant differences in transcript expression between human and chimpanzee.
Our data are consistent with a retroviral infection that bombarded the genomes of chimpanzees and gorillas independently and concurrently, 3–4 million years ago. We speculate on the potential impact of such recent events on the evolution of humans and great apes.
Indeed, the paper on the sequencing of the chimpanzee genome I cited above, cites amongst its findings that the chimpanzee and gorilla lineages acquired new families of retroviral insertions
after the chimpanzee/gorilla and human lineages diverged. The above paper merely adds support to that finding. And, lo and behold, the new additions in the chimpanzee/gorilla lineage took place
before those two lineages diverged, indicating that the common ancestor of humans and other great apes gave rise to two lineages, one resulting in humans, the other resulting in a common ancestor for chimpanzees and gorillas.
I think that's game over somehow.
But just as you thought you were coming to the end of this, this is just Part 1. I'll post Part 2 in a moment.










